Grammatical Evolution Tutorial:
This tutorial presents a short, easy problem to solve using Grammatical Evolution techniques with this software. The example uses only the grammatical evolution portion rather than a hybrid for clarity.
Installation
The quickest way to install is with easy_install. Since this is a Python library, at the Python prompt put:
easy_install pyneurgen
The Problem
The problem is fairly trivial, but should be sufficient to gain an appreciation for methods to solve problems using grammatical evolution. The problem to solve:
For values 0 to 99, what expression could be used to minimize:
abs(expression - pow(x, 3))
The program will come up with an appropriate answer.
The first step, import modules that we will use:
from pyneurgen.grammatical_evolution import GrammaticalEvolution from pyneurgen.fitness import FitnessElites, FitnessTournament from pyneurgen.fitness import ReplacementTournament, MAX, MIN, CENTER
The next task is to specify a grammar that will be referenced as well as a stub of a program, or at least an appropriate starting point to get the evolutionary process running. Then, we define some of the starting parameters.
Backus-Naur form
The point of the grammar is to define variables and the possible assignments that can be made to the variables. It is by the use of grammatical evolution that assignments of the variables will be made, and final assembly of a program. There is one program generated per genotype. Any language can be defined by the grammar, as long as there is an appropriate executor when it comes to runtime of that program. In this case we will stick with Python. The grammar uses a Backus-Naur form (BNF), with a structure of:
<variable> ::= this | is | a | list | of | options
A variable is identifiable because of the angle brackets around it. It is also separated from the list of options by ::=. The following is the BNF that will be used for this example:
bnf = """ <expr> ::= <expr> <biop> <expr> | <uop> <expr> | <real> | math.log(abs(<expr>)) | <pow> | math.sin(<expr> )| value | (<expr>) <biop> ::= + | - | * | / <uop> ::= + | - <pow> ::= pow(<expr>, <real>) <plus> ::= + <minus> ::= - <real> ::= <int-const>.<int-const> <int-const> ::= <int-const> | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0 <S> ::= import math total = 0.0 for i in xrange(100): value = float(i) / float(100) total += abs(<expr> - pow(value, 3)) fitness = total self.set_bnf_variable('<fitness>', fitness) """
As you can see, there are a number of variables and optional values that can take place. Note that some of the options are variables that have angled brackets, and some do not. Those that do not are terminals; the selection process ends when a terminal is selected. If a variable selected, the program uses the BNF to recursively evaluate the value of the variables found until a terminal is reached.
For example, suppose that <real> is selected. The program will see <int-const>.<int-const> and will evaluate the variables one by one. Upon looking up the definition of <int-const>, it will find a list that could be any of the following elements:
<int-const> | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 0]
Using a selection process, which will be covered below, the program will select a number or the <int-const> and recursively operate until <real> is defined as a number. At that point, the program will continue with the next variable.
There is also the <S> which is the starting point. The starting point can be minimal and open-ended, or fairly structured and limited in scope, depending upon the needs of the problem. One other thing, when laying out the starting point in code, take into account the use of whitespace in Python. Since this BNF is defined using a string in the form of """text""", the returns and spacing will be used in formatting the statements. That is the explanation for the unaesthetic yet important layout above. This caveat only applies to a series of statements in the BNF, not the definitions of variables. There are examples found in the demos directory with this package, if there is still some confusion about it.
Implementation
There are some features here that are specific to this implementation.
Note that from a Python standpoint that it has self. in the front of the fitness desnignator. That is because the fitness computation takes place within the Genotype namespace. If for some reason, you would like to intoduce a function from the Genotype class, it can be available in your program.
Another implementation specific feature is self.set_bnf_variable('<value>', value). The purpose is to save a variable within the BNF during runtime. This can be useful to temporarily assign during the course of the program. This feature enables the possibility of extending the BNF during runtime.
You should be careful, however, when saving variables that the spirit of reproducability is maintained. While assignment of variables is apparently random, you will want a specific genotype to always assign in exactly the same fashion.
Finally, there is an implementation specific feature that must be followed. When the fitness computation has ended, the <fitness> variable with the BNF is used as the fitness value. In this case, total is assigned to fitness, which is then saved to the BNF. The sheer redundancy is used here to help emphasize that if there is no fitness assigned upon completion all of your genotypes will fail every time.
Mapping from the Genotype
It is the structure of the genotype that defines the choices that are made for options. Genotypes in genetic algorithms, and with this implementation as well, use strings such as '01010111' and '10110111' to define a genotype. Grammatical evolution translates those values 8 bits at time into lists of decimal values, such [73, 45, 56, 1, 101]. It is this list that will be used as the basis for determining what option is selected.
The basis for selection derives from the use of remainders. The following example will make this clear: Suppose that <biop> has been selected and the mapping process needs to determine whether the choice is '+', '-', '*', or '/'. The mapping process takes the next number from the genotype in the list. Suppose that number is 73. The choice is then made on the basis of:
options = ['+', '-', '*', '/'] codon = 73 choice_number = codon % len(options) choice = options[choice_number]
This results in a choice_number of 1, and the '-' option selected.
That choice made, the next substitution will be made on the basis of the next number in the genotype sequence. When all the numbers in the list have been drawn, then the genotype sequence can wrap around and start at the beginning. The program defaults to wrapping around. If the end has been reached, yet more are needed, then the genotype will be marked as a failure and move on to the next. The genotype can also be extended in lengh with the wrapped values as well. The rationale for this is that mutations in an extended genotype might be able to make minor changes in the extended genotype without affecting the earlier portions. It is not totally clear that this arrangement is any better than simply starting out with a longer genotype.
Setting the Evolutionary Parameters
Genotype Lengths
To review, the modules have been imported, and the BNF has been defined. Now the parameters for running the program need to be set. Below, the GrammaticalEvolution class is substantiated, and an initial length is set at 20, extending up to 500. This length, by the way, is the length of the decimal list of numbers. The population size is set to 50, and the genotype will wrap if it hits the end of the genotype list. While mapping the genotype to the fitness program uses the decimal form of the genotype, crossovers and mutations are performed on the binary form.
Also shown below is loading the BNF into the GrammaticalEvolution class. The load converts it to a dict form with each variable name as a key.
ges = GrammaticalEvolution() ges.set_bnf(bnf) ges.set_genotype_length(start_gene_length=20, max_gene_length=50) ges.set_population_size(50) ges.set_wrap(True)
Stopping Criteria
The next set of parameters control the course of the process. The set_max_generations function sets the maximum number of generations that will be run. If a fitness value is also set, the number of process will halt upon reaching the goal. If the goal is not reached, then it will stop at the maximum generations.
ges.set_max_generations(1000) ges.set_fitness_type(MIN, .01)
The fitness selection process for each generation in this case is to minimize the difference between the generated number and the cube of the number. In other words, the closer the fitness value is to zero, the more fit that it is likely to be. Fitness types have a choice of 'max', 'min', 'center'. Given that we want to hit zero, the fitness_type in this case is 'center', and if a fitness value gets below .01, then that is deemed close enough.
Genotype Failures
Declaring failure is an important part of this process. Failure declaration can occur for a number of reasons:
- The program generated grows to greater than 500 characters
- The program generated is syntactically incorrect
- It takes more than 10 seconds to generate the program.
- It takes more than 120 seconds to run the program.
- Some program fault such as dividing by zero.
When failure has occurred, the fitness value is set to the fitness_fail value. Since every problem can be a little different, in this case the fail value is set 100.0, since we want something close to zero. That way, the likelihood of replacement of that genotype is drastically increased.
ges.set_max_program_length(500) ges.set_timeouts(10, 120) ges.set_fitness_fail(100.0)
Fitness Selections
The selection process for fitness should emphasize selecting successful genotypes and replacing less successful versions. However, success being a relative thing, including some less successful versions in a later generation might well lead through crossovers and mutations to a much more successful genotype. In this program, up to 50% are being replaced. We are using two types of fitness selection, FitnessElites, and FitnessTournament. FitnessElites takes the top 5% in this case and puts them in the list for reproduction and mutation. Then, a tournament process is used, where randomly, a pair of genotypes are drawn with replacement and the best genotype is added to the list. This continues until the fitness quota is met, 50% in this case.
ges.set_fitness_selections( FitnessElites(ges.fitness_list, .05), FitnessTournament(ges.fitness_list, tournament_size=2)) ges.set_max_fitness_rate(.5)
Once the fitness selection has been made, genotypes are modified by both the crossover operator and the mutation operator. Both operators act on the binary version of the genotype. Crossovers are performed on 20% in this case. Crossovers take place by selecting pairs, swapping from a single point parts of binary genotypes. A choice can be made for the number of children, either one or two. If one is selected, that one of the two crossed is selected randomly. In this case we are using two.
The mutation operator can act in one of two ways. It can select a genotype for mutation at the mutation probability rate. If mutated, it changes one bit in the genotype. This is a mutation type of 's' for single. Or, each genotype in the fitness selection list can be mutated at the probabilistic mutation rate on a bit by bit basis. This mutation type is 'm' for multiple, since a particular genotype is at risk to be modified multiple times, depending upon the mutation rate. In this case, 'multiple' has been selected and the mutation rate is 2.5%
ges.set_mutation_rate(.025) ges.set_fitness_selections( FitnessElites(ges.fitness_list, .05), FitnessTournament(ges.fitness_list, tournament_size=2)) ges.set_max_fitness_rate(.5) ges.set_crossover_rate(.2) ges.set_children_per_crossover(2) ges.set_mutation_type('m') ges.set_max_fitness_rate(.25)
Replacements
The replacement process is very similar to the fitness selection, although in reverse. In this case, ReplacementTournament is used where a tournament is conducted with 3 participants, and the worst is replaced. Replacements continue until all of the genotypes on the fitness selection list have been replaced.
ges.set_replacement_selections( ReplacementTournament(ges.fitness_list, tournament_size=3))
Other
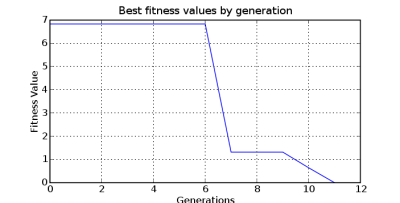
A history of fitness lists can be maintained through a run. Seeing the history of fitness values per generation can give much insight into other ways to evaluate the problem. Summary values from each list are available as well, such as mean, median, best_value, worst_value, and standard deviations.
ges.set_maintain_history(True)
Run
With all the parameters set, the only thing left to do is create the genotypes and run.
ges.create_genotypes() print ges.run()
Results
On a sample run the best genotype member was 30. The fitness_list structure is in the format:
[[fitness value, member number1], [[fitness value, member number2]]
The following is ges.fitness_list printed in raw form, sorted by best value
[[8.6717539074000001e-16, 30], # This one hit the mark [2.7773611289, 24], [6.8311913649699996, 0], # These get progressively worse [6.8311913649699996, 6], [6.8311913649699996, 21], [6.8311913649699996, 28], [6.8311913649699996, 34], [6.8311913649699996, 41], [7.2573146880300001, 12], [7.8597593677799997, 42], [7.8597593677799997, 47], [8.3324999999999996, 10], [8.3324999999999996, 11], [8.3324999999999996, 13], [8.3324999999999996, 15], [8.3324999999999996, 27], [8.3324999999999996, 33], [8.3324999999999996, 35], [8.3324999999999996, 36], [8.3324999999999996, 37], [8.3324999999999996, 43], [8.3324999999999996, 44], [8.3324999999999996, 48], [10.3730644307, 5], [14.7910856822, 45], [15.7354593807, 7], [23.036235317300001, 46], [100.0, 1], # Program length too long or [100.0, 2], # invalid for some other reason [100.0, 3], [100.0, 4], [100.0, 8], [100.0, 9], [100.0, 14], [100.0, 16], [100.0, 17], [100.0, 18], [100.0, 19], [100.0, 22], [100.0, 23], [100.0, 25], [100.0, 26], [100.0, 29], [100.0, 31], [100.0, 32], [100.0, 38], [100.0, 39], [100.0, 40], [100.0, 49], [1754878323.3599999, 20]] # Legal program, really bad values
The generated program is:
import math total = 0.0 for i in xrange(100): value = float(i) / float(100) total += abs(pow(value, 3.0) - pow(value, 3)) fitness = total self.set_bnf_variable('<fitness>', fitness)